Map
The Map function takes a reference file, an output filename,
the chromosome strand on which the operation should be applied, and an
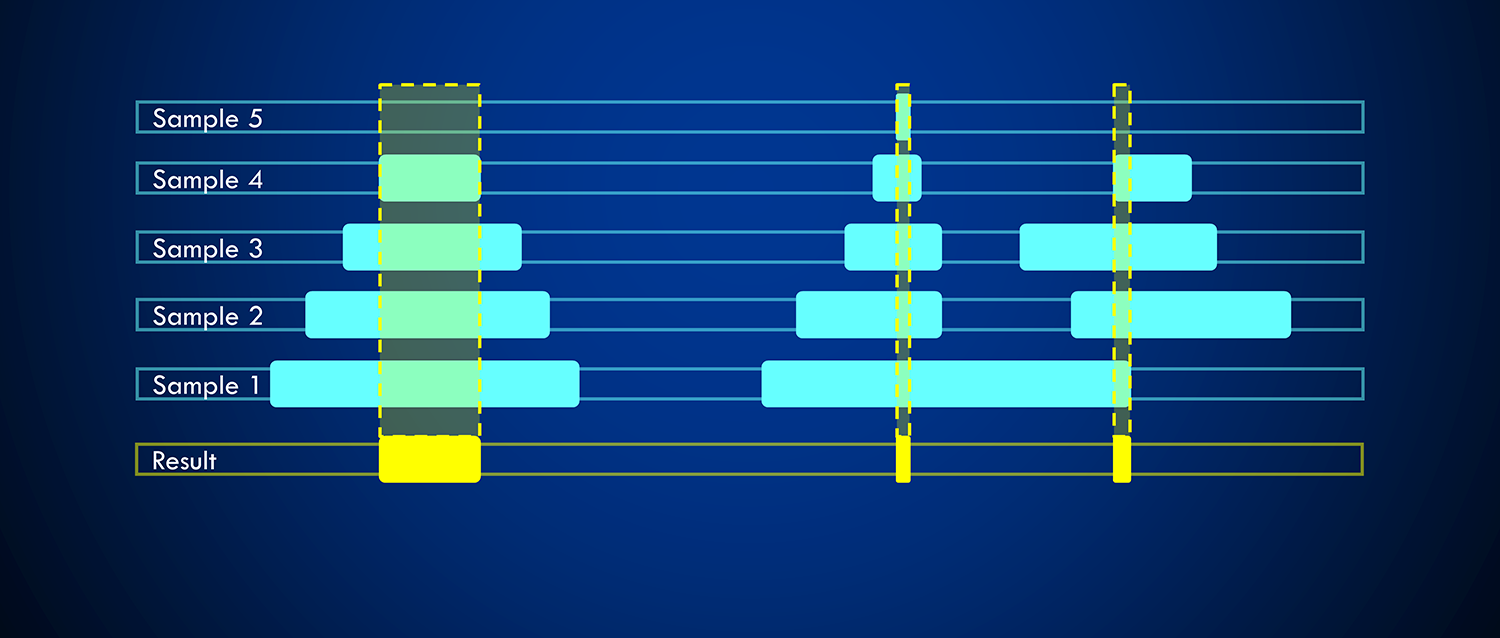
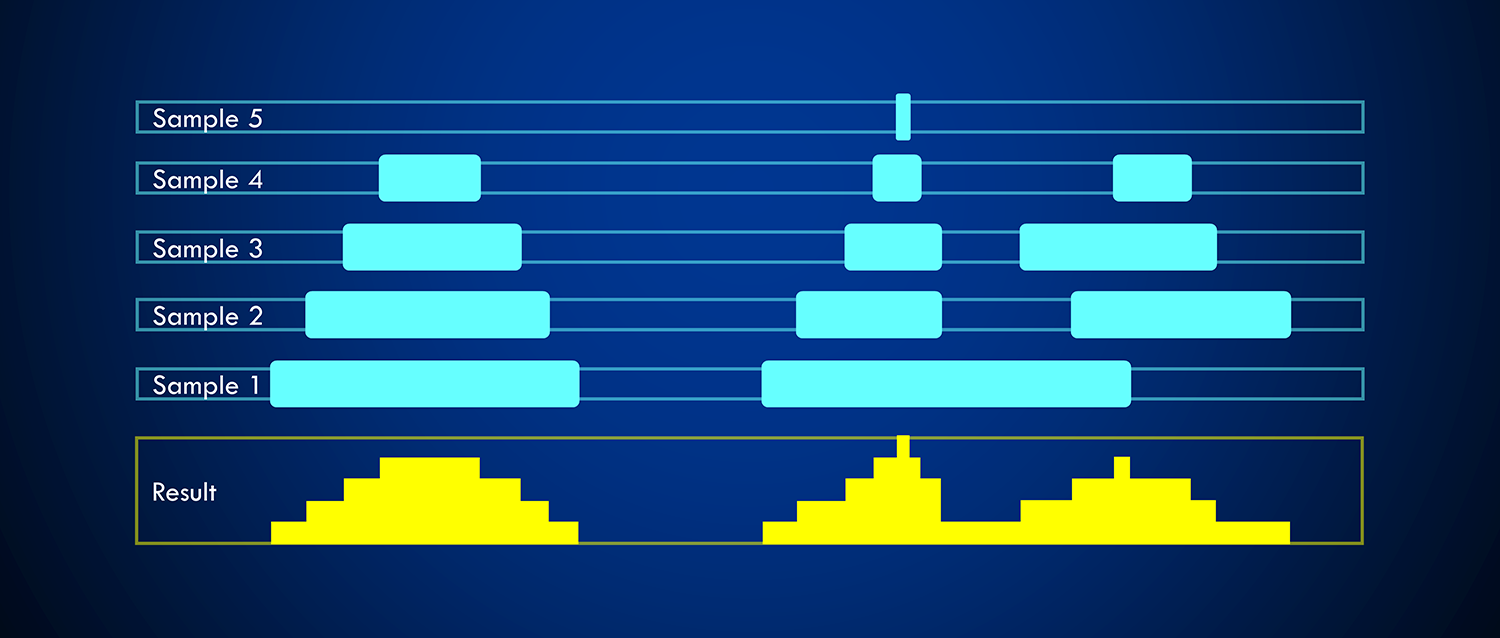
aggregation function. See the following example, where the highlighted intervals
from the samples are determined as intervals overlapping with the given reference
interval.

The Map function is exposed via Di4BCLI (the command line
interface to Di4's bioinformatics wrapper). The Di4BCLI defines the following
syntax for the Mapfunction:
Map reference results_file strand aggregate_function
For instance:
Map C:\Users\Vahid\Desktop\Di4Data\ref.narrowpeak res.bed * count
Where * means “un-stranded” as the provided input data
do not define strand. count is an aggregation function,
which counts the number of intervals overlapping a reference interval.
As a result of executing this command, Di4BCLI shows the following message.
> Map C:\Users\Vahid\Desktop\Di4Data\ref.narrowpeak res.bed * count
Loaded #i: 196,180 ET: 00:00:01.0622689 Speed: 184,680 #i\sec
Map #i: 196,180 ET: 00:00:00.2214104 Speed: 886,047 #i\sec
Export #i: 196,111 ET: 00:00:00.1353136 Speed: 1,449,307 #i\sec
-: Done ... Overall ET: 00:00:01.4325864
Regarding the runtime; Di4’s runtime is the runtime of the Map function,
which is 00:00:00.2214104 in this run. The load and export
times are Di4B and Di4BCLI runtime for loading the reference
sample, calling independent instances of Di4 instances (Chr-level degree
of parallelism), and saving the results.